Setting up Local Swiss-Prot Database and BLAST Searching

NCBI provides a web interface to run BLAST searches. However, it is also possible to download the databases to search locally.
This post shares the process of downloading the BLAST+ suite of command-line tools, setting up the Swiss-Prot database, and performing a simple blastp search.
The source code is available on GitHub here.
BLAST+ Installation
BLAST+ is a suite of command-line tools to manage BLAST nucleotide and protein databases and perform BLAST searches. It can be downloaded from the NCBI's website here.
The downloadable used in this post is ncbi-blast-2.16.0+-aarch64.dmg, where ncib-blast is the name, 2.16.0+ is the version, aarch64 refers to the hardware architecture (common to modern Macs with Apple Silicon), and .dmg specifies Apple Disk Image file format (program installer).
The BLAST+ installation encountered two challenges.
First, the app triggers a security warning:
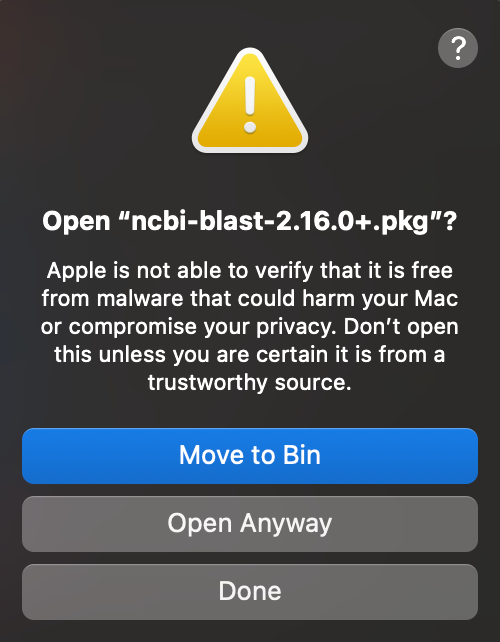
This happens because the installer has not been signed with Apple Developer ID certificate and then notarised. The app still works but the warning does detract from an otherwise straightforward installation.
The second challenge is the need to manually add the BLAST+'s location to PATH, which is necessary to be able to call BLAST+ tool from any directory. The default installation location is /usr/local/ncbi/blast. Here is the list of programs expected to be found at that location:
> ls -lh /usr/local/ncbi/blast/bin
total 1115896
-rwxr-xr-x 1 root wheel 26M 26 Jun 2024 blast_formatter
-rwxr-xr-x 1 root wheel 30M 26 Jun 2024 blast_formatter_vdb
-rwxr-xr-x 1 root wheel 19M 26 Jun 2024 blast_vdb_cmd
-rwxr-xr-x 1 root wheel 16M 26 Jun 2024 blastdb_aliastool
-rwxr-xr-x 1 root wheel 17M 26 Jun 2024 blastdbcheck
-rwxr-xr-x 1 root wheel 23M 26 Jun 2024 blastdbcmd
-rwxr-xr-x 1 root wheel 26M 26 Jun 2024 blastn
-rwxr-xr-x 1 root wheel 30M 26 Jun 2024 blastn_vdb
-rwxr-xr-x 1 root wheel 26M 26 Jun 2024 blastp
-rwxr-xr-x 1 root wheel 26M 26 Jun 2024 blastx
-rwxr-xr-x 1 root wheel 6.1K 8 Aug 2019 cleanup-blastdb-volumes.py
-rwxr-xr-x 1 root wheel 17M 26 Jun 2024 convert2blastmask
-rwxr-xr-x 1 root wheel 26M 26 Jun 2024 deltablast
-rwxr-xr-x 1 root wheel 16M 26 Jun 2024 dustmasker
-rwxr-xr-x 1 root wheel 4.6K 13 May 2021 get_species_taxids.sh
-rwxr-xr-x 1 root wheel 50K 27 May 2020 legacy_blast.pl
-rwxr-xr-x 1 root wheel 18M 26 Jun 2024 makeblastdb
-rwxr-xr-x 1 root wheel 17M 26 Jun 2024 makembindex
-rwxr-xr-x 1 root wheel 18M 26 Jun 2024 makeprofiledb
-rwxr-xr-x 1 root wheel 26M 26 Jun 2024 psiblast
-rwxr-xr-x 1 root wheel 26M 26 Jun 2024 rpsblast
-rwxr-xr-x 1 root wheel 26M 26 Jun 2024 rpstblastn
-rwxr-xr-x 1 root wheel 16M 26 Jun 2024 segmasker
-rwxr-xr-x 1 root wheel 26M 26 Jun 2024 tblastn
-rwxr-xr-x 1 root wheel 30M 26 Jun 2024 tblastn_vdb
-rwxr-xr-x 1 root wheel 26M 26 Jun 2024 tblastx
-rwxr-xr-x 1 root wheel 36K 17 Apr 2024 update_blastdb.pl
-rwxr-xr-x 1 root wheel 20M 26 Jun 2024 windowmasker
This post explores blastp, update_blastdb.pl, and blastdbcmd commands. The output also shows that the last modification date of any of the files is June 2024, which, as of July 2025, means that there have been no updates to BLAST+ in over a year.
The location can be added to PATH by running the following command:
export PATH="/usr/local/ncbi/blast/bin:$PATH"
Finally, the installation can be verified by checking the tool version. Here is the expected output:
> blastp -version
blastp: 2.16.0+
Package: blast 2.16.0, build Jun 25 2024 08:57:39
Other options can be found by running the help command: blastp -help.
Swiss-Prot Download
The first choice to make is which database to download. In this case, the goal is just to experiment with the download and search process. As such, any database will do. So swissprot, as one of the most lightweight databases, is chosen.
The full list of available databases in alphabet order can be checked with the following command:
> update_blastdb.pl --showall | sort | nl
1 16S_ribosomal_RNA
2 18S_fungal_sequences
3 28S_fungal_sequences
4 Betacoronavirus
5 core_nt
6 env_nr
7 env_nt
8 human_genome
9 ITS_eukaryote_sequences
10 ITS_RefSeq_Fungi
11 landmark
12 LSU_eukaryote_rRNA
13 LSU_prokaryote_rRNA
14 mito
15 mouse_genome
16 nr
17 nt
18 nt_euk
19 nt_others
20 nt_prok
21 nt_viruses
22 pataa
23 patnt
24 pdbaa
25 pdbnt
26 ref_euk_rep_genomes
27 ref_prok_rep_genomes
28 ref_viroids_rep_genomes
29 ref_viruses_rep_genomes
30 refseq_protein
31 refseq_rna
32 refseq_select_prot
33 refseq_select_rna
34 SSU_eukaryote_rRNA
35 swissprot
36 taxdb
37 tsa_nr
38 tsa_nt
The second choice to make is selecting download location, which is specified by the BLASTDB environment variable. One reasonable approach involves one global location to use for all databases and all projects. If a project has peculiarities requiring a different database, it can override BLASTDB environment variable to a different location.
The first commit sets up a shell script to configure the BLASTDB environment variable and download the swissprot database:
# Configure path to blast databases
if [ -z "$BLASTDB" ]; then
export BLASTDB=~/databases/blast/
{
echo ""
echo "# Set path to BLAST databases"
echo "export BLASTDB=\$HOME/databases/blast/"
} >> ~/.zprofile
source ~/.zprofile
fi
# Download swissprot
mkdir -p $BLASTDB
cd $BLASTDB
update_blastdb.pl --decompress swissprot
The above script also adds BLASTDB to the shell startup file (.zprofile), which runs whenever a shell launches. As a result, BLASTDB will be automatically set in any new terminal sessions, ensuring that it does not need to be set again.
One downside of the above script is the risk of adding duplicate lines to .zprofile. After the very first run, the .zprofile file is updated but the BLASTDB variable is still empty in the current session. So re-running the script updates the .zprofile file again. The current session has to be restarted to avoid this problem.
Another potential issue is that depending on how the init.sh file was created, it may lack executable permissions. The command chmod +x init.sh adds them.
Once downloaded, the results can be checked with the following command:
> blastdbcmd -info -db swissprot
Database: Non-redundant UniProtKB/SwissProt sequences
485,565 sequences; 184,945,355 total residues
Date: Jul 1, 2025 4:45 AM Longest sequence: 35,213 residues
BLASTDB Version: 5
Volumes:
~/databases/blast/swissprot
Example Usage
The second commit tries an example blastp search, which successfully finds a number of matches:
blastp \
-query query.fasta \
-db swissprot \
-out results.txt \
-outfmt 6
The inputs are specified in the query.fasta file., while the outputs are sent to the results.txt file in the output format number 6. In this case the search returned 20 matches:
> wc -l results.txt
20 results.txt
> head -n 1 results.txt
example_protein B1K1H7.1 100.000 40 0 0 1 40 189 228 1.66e-20 84.7
According to the documentation (accessible via blastp -help command), the output format number 6 is tabular with the following column names:
- qaccver (Query accession.version)
- saccver (Subject accession.version)
- pident (Percentage of identical matches)
- length (Alignment length)
- mismatch (Number of mismatches)
- gapopen (Number of gap openings)
- qstart (Start of alignment in query)
- qend (End of alignment in query)
- sstart (Start of alignment in subject)
- send (End of alignment in subject)
- evalue (Expect value)
- bitscore (Bit score)